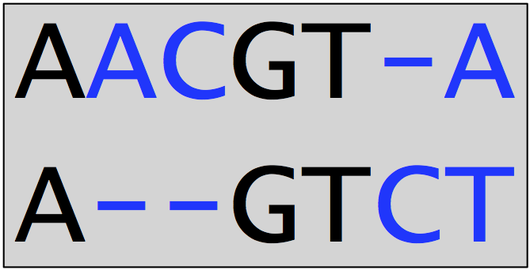Edit alignment score is an alignment scoring function that assigns a (negative) unit cost to every edit operation appearing in the alignment and a cost of 0 to every matched symbol. Note that every edit operation in the alignment corresponds to a mismatched symbol (where a mismatched gap symbol corresponds to an insertion or deletion).
As a result, if
A very small alignment of DNA strings is shown below. Note that this alignment implies two deletions, one insertion, and one substituted nucleotide. There are 4 total positions at which symbols do not match, so that the edit score of the alignment is -4.