Aug. 23, 2012, midnight by btjaden
Topics: Combinatorics, Dynamic Programming, Graph Algorithms
Don't Look Down
We have discussed the problem of counting RNA secondary structures in previous problems. In this problem, we will add some assumptions to those used in “Motzkin Numbers and RNA Secondary Structures” to provide ourselves with an ultimately robust way of counting feasible RNA secondary structures.
First, in addition to the classic Watson and Crick base pairing of adenine with uracil and cytosine with guanine, uracil sometimes bonds with guanine, forming what is called a wobble base pair. As a result, we would like to allow wobble base pairing.
Second, although RNA folds over itself during base pairing, the structure of the molecule is rigid enough to prevent bases located very close to each other on the strand from bonding to each other.
Given an RNA string
We say that a matching in the bonding graph for
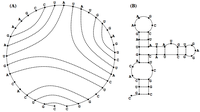
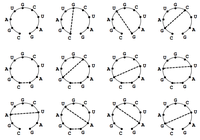
See Figure 1 for an example of a valid matching if we allow wobble base pairs. In this problem, we will wish to count all possible valid matchings in a given bonding graph; see Figure 2 for all possible valid matchings in a small bonding graph, assuming that we allow wobble base pairing.
Given: An RNA string
Return: The total number of distinct valid matchings of basepair edges in the bonding graph of
AUGCUAGUACGGAGCGAGUCUAGCGAGCGAUGUCGUGAGUACUAUAUAUGCGCAUAAGCCACGU
284850219977421