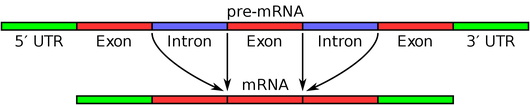
RNA splicing is the process by which a molecule of pre-mRNA (typically corresponding to a gene in DNA) is modified to remove regions that will not be translated into protein.
Specifically, the pre-mRNA is composed of introns alternating with exons. During RNA splicing, the introns are removed to create a new strand of RNA for protein translation; see the figure below for a simple illustration.
RNA splicing is facilitated by a spliceosome, a macromolecule formed of a mixture of RNA and proteins. The simplest analogy for how RNA splicing is physically achieved is by imagining a long string of putty. For a given intron, a loop is formed, and the neighboring exons are squeezed together, in the process pinching off the loop containing the intron.
A wider diversity of proteins can be created in the process of alternative splicing. Several different forms of alternative splicing have been observed, but the most common form is exon skipping, in which a number of exons are deleted along with the introns.