A restriction enzyme, also known as a restriction endonuclease, is an enzyme created by bacteria to counteract phages. The enzyme is programmed with a "recognition sequence," a short interval of contiguous DNA nucleotides that it can bind to, cutting the DNA strand somewhere in the middle of the interval.
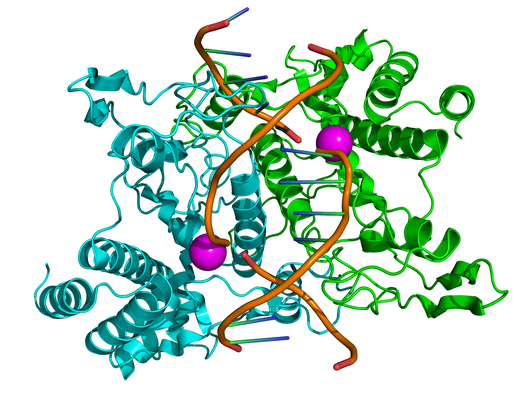
In order to successfully disarm viral DNA, a restriction enzyme must cleave through both strands of the double helix. For this reason, restriction enzymes are homodimers, meaning that they are composed of two identical substructures, each of which carries out the task of cutting DNA with the same recognition sequence (see the figure for an illustration of Type II site-specific deoxyribonuclease, with its two identical substructures colored green and blue).
The issue, however, is that if the same recognition sequence is located in two locations greatly separated on the same molecule of DNA, then the likelihood that the restriction enzyme will cleave both strands of DNA decreases. In fact, its likelihood of success is maximized when the two recognition sequences are located directly across from each other on opposing strands, which occurs precisely when the recognition sequence is a reverse palindrome, or a string that is equal to its reverse complement. As a result, the recognition sequence must have even length; most are 6 nucleotides long, in order to increase the probability that the recognition sequence will be located.
Bacteria also must protect against restriction enzymes cutting their own DNA. To this end,
they employ a process called DNA methylation, in which they add to their own DNA methyl groups (