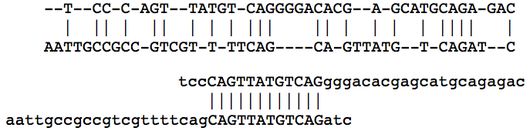
A local alignment of two strings is an alignment of a substring of one of the strings with a substring of the other. Local alignment is in contrast with a global alignment, in which every symbol of both strings is aligned; as a result, in local alignment, only those symbols belonging to the substrings that are aligned will contribute to the calculation of the alignment's score. This serves a practical purpose when we wish to search large genetic strings for small regions of the highest similarity.
In the figure below, an optimal global alignment of two DNA strings is shown, followed by an optimal local alignment of the same two strings. The edit alignment score function is used in both cases; the local alignment of the two strings has a net score equal to 0 because the symbols outside the matched interval are not counted. This figure demonstrates the importance of local alignment in applications, in which we often search very long genetic strings for small, local areas of similarity (e.g., finding shared genes along an interval of two chromosomes).